1.3. Systems of ODEs#
So far we have only looked at the solution of a single ODE. In most practical cases, a system of multiple ODEs is required to model a real-world situation. Recall that a single first-order ODE is expressed as a function of the single independent variable \(t\) and a dependent function \(y\)
A system of first-order ODEs is expressed as a set of multiple ODEs such that
where \(y_1(t), y_2(t), \ldots, y_N(t)\) are multiple dependent functions. An IVP involving a system of ODEs requires an initial value for each equation in the system, i.e., \(y_1(t_0) = a_1\), \(y_2(t_0) = a_2\) etc. where \(a_1, a_2, \ldots, a_N\) are constants.
To apply a numerical method to solve a system of ODEs, we can write it in vector form \(\mathbf{y}' = f(t, \mathbf{y})\) where
Recall that the Euler method for solving an IVP for a single ODE is
then for solving a system of first-order ODEs we simply replace \(y_n\) and \(f(t_n, y_n)\) with \(\mathbf{y}_n\) and \(f(t_n, \mathbf{y}_n)\)
Example 1.2
The SIR model is a simple model that describes the spread of an infectious disease in a population. The model divides the population into three compartments: those who have not yet contracted the disease but are susceptible (\(S\)) those who are infected (\(I\)) and those who have recovered and gained immunity (\(R\)). The formulation of the model is
where \(N = S + I + R\) is the total population, \(\beta\) is the infection rate in the number of people per day who become infected, and \(\gamma\) is the recovery rate at which a person who is infected recovers per day.
A disease breaks out in a population where \(\beta = 0.5\), \(\gamma = 0.1\) and \(S(0) = 99\), \(I(0) = 1\) and \(R(0) = 0\). Calculate the solution to the SIR model over the first 50 days using the Euler method with a step length of \(h = 1\).
Solution
Let \(y_1 = S\), \(y_2 = I\) and \(y_3 = R\) and writing the SIR model in vector form we have
The initial conditions are \(\mathbf{y}_0 = (99, 1, 0)\) and \(N = 100\), \(\beta = 0.5\), \(\gamma = 0.1\) and \(\frac{\beta}{N} = 0.005\). Calculating the first few steps of the Euler method with \(h = 1\)
The plot of the solutions to the SIR model using the Euler method for the first 50 days of the infection is shown in Fig. 1.9
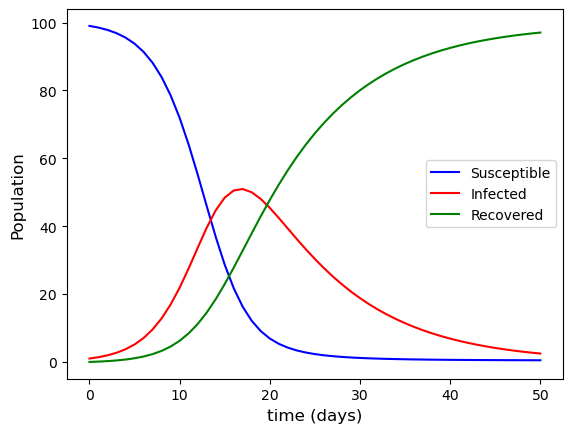
Fig. 1.9 Euler method solutions for the SIR model with \(S(0) = 99\), \(I(0) = 1\), \(R(0) = 0\), \(\beta = 0.5\) and \(\gamma = 1\).#
1.3.1. Code#
The function euler() which computes the solutions to a system of ODEs using the Euler method is shown below. This is very similar to the function used to solve a single ODE with the exception that y is initialised to be a \((nsteps + 1) \times m\) array where \(m\) is the number of equations in the system. The first row of y is set equal to the initial values y0 and a for loop is used to step through the Euler method.
def euler(f, tspan, y0, h):
m = len(y0)
nsteps = int((tspan[1] - tspan[0]) / h)
t = np.zeros(nsteps + 1)
y = np.zeros((nsteps + 1, m))
t[0] = tspan[0]
y[0,:] = y0
for n in range(nsteps):
y[n+1,:] = y[n,:] + h * f(t[n], y[n,:])
t[n+1] = t[n] + h
return t, y
function [t, y] = euler(f, tspan, y0, h)
nsteps = floor((tspan(2) - tspan(1)) / h);
m = length(y0);
t = zeros(1, nsteps + 1);
y = zeros(nsteps + 1, m);
t(1) = tspan(1);
y(1,:) = y0;
for n = 1 : length(t) - 1
y(n+1,:) = y(n,:) + h * f(t(n), y(n,:));
t(n+1) = t(n) + h;
end
y = y;
end
Note that this function assumes that the ODE function returns an \(N \times 1\) vector.
The code below sets up and solves the SIR model example from Example 1.2 using the Euler method
import numpy as np
import matplotlib.pyplot as plt
# Define Euler method function
def euler(f, tspan, y0, h):
m = len(y0)
nsteps = int((tspan[1] - tspan[0]) / h)
t = np.zeros(nsteps + 1)
y = np.zeros((nsteps + 1, m))
t[0] = tspan[0]
y[0,:] = y0
for n in range(nsteps):
y[n+1,:] = y[n,:] + h * f(t[n], y[n,:])
t[n+1] = t[n] + h
return t, y
# Define SIR model
def SIR(_, y):
S, I, R = y
N = S + I + R
dS = -beta / N * I * S
dI = beta / N * I * S - gamma * I
dR = gamma * I
return np.array([ dS, dI, dR ])
# Define IVP parameters
tspan = [0, 50] # boundaries of the t domain
y0 = [99, 1, 0] # initial values
beta, gamma = 0.5, 0.1 # Model parameters
h = 1 # step length
# Solve the IVP using the Euler method
t, y = euler(SIR, tspan, y0, h)
# Plot solution
fig, ax = plt.subplots()
plt.plot(t, y[:,0], "b-", label="Susceptible")
plt.plot(t, y[:,1], "r-", label="Infected")
plt.plot(t, y[:,2], "g-", label="Recovered")
plt.xlabel("time (days)")
plt.ylabel("Population")
plt.legend()
plt.show()
% Define IVP parameters
tspan = [0, 50]; % boundaries of the t domain
y0 = [99, 1, 0]'; % initial values [S, I, R]
beta = 0.5; % infection rate
gamma = 0.1; % recovery rate
h = 1; % step length
% Solve the IVP using the Euler method
[t, y] = euler(@(t, y)SIR(t, y, beta, gamma), tspan, y0, h);
% Plot solution
hold on
plot(t, y(:,1), 'b-', LineWidth=2)
plot(t, y(:,2), 'r-', LineWidth=2)
plot(t, y(:,3), 'g-', LineWidth=2)
hold off
xlabel('$t$', Fontsize=14, Interpreter='latex')
ylabel('$y$', Fontsize=14, Interpreter='latex')
legend('Susceptible', 'Infected', 'Recovered', Location="east")
axis padded
% ---------------------------------------------------------------
function [t, y] = euler(f, tspan, y0, h)
nsteps = floor((tspan(2) - tspan(1)) / h);
m = length(y0);
t = zeros(1, nsteps + 1);
y = zeros(nsteps + 1, m);
t(1) = tspan(1);
y(1,:) = y0;
for n = 1 : length(t) - 1
y(n+1,:) = y(n,:) + h * f(t(n), y(n,:));
t(n+1) = t(n) + h;
end
end
function y = SIR(~, y, beta, gamma)
S = y(1);
I = y(2);
R = y(3);
N = S + I + R;
y = [ -beta / N * I * S ,
beta / N * I * S - gamma * I,
gamma * I ];
end
Note
Note that the SIR model has been defined using a non-autonomous function so is placed at the bottom of the script file. The euler() function takes in 2 inputs t and y, so the addition input parameters beta and gamma are passed in using @(t, y)SIR(t, y, beta, gamma).